Learn how to convert EDF files produced by medical devices to HFiles, and use it efficiently with Warp 10 to perform performant analytics.

Modern medical devices produce health data and often at high frequency. Those data are read-only of course and can represent a huge amount of measurements. Some devices store it in binary compressed files locally.
We will show you how to handle that kind of file with Warp 10, thanks to HFiles.
EDF file format
Many medical devices use the EDF file format (short for European Data Format) to store measurements. This kind of file was specified in 1992 and is still in use. This is a binary file format in which you can store signals and annotations (since the EDF+ version).
A file could contain one or more signals (measurements) and one or more recordset per signal. The only existing timestamp is the record start date. For each measure, you must compute the timestamp with the record position and the sampling frequency.
For example, if we model that kind of file in JSON format:
[
{
"name': "Signal A",
"start-date" : "2022-02-14 18:55:45",
"frequency": 0.5,
"records": [
[ 0: 1, 1: 1.2, 2: 0.3 ],
[ 3: 0.5, 4: 1.9, 5: 0.5 ]
]
}
]It means that there is 0.5 sample per second. So, if we translate that into a real-time series we should have something like that (of course, this is not a valid GTS representation):
{
"name": "Signal-A"
"values": [
"2022-02-14 18:55:45": 1
"2022-02-14 18:55:47": 1.2
"2022-02-14 18:55:49": 0.3
"2022-02-14 18:55:51": 0.5
...
]
}Annotations are quite differently handled. The main format is:
[
{
"name": "Annotation A"
"records": [
[ 0::"event a", 10:5:"event b", <time offset>:[optional duration]:<event name> ],
...
]
}
]It exists some libs in Python, Java, Javascript, or C which parse this kind of file, but some of them do not handle both EDF and EDF+ files and could be a little buggy.
Practice
For this example, we will use a CPAP (Continuous Positive Airway Pressure) system dedicated to sleep apnea treatment.

(by MyUpchar, Wikimedia Commons CC BY-SA 4.0)
This kind of system often produces EDF files (one per sleep session) with a lot of measurements:
- Tidal volume
- Mask pressure
- Pump pressure
- Mask air leaks
- Snore
- Respiration rate
- Flow
- events like Apnea, Hypopnea, Obstructive Apnea, Central Apnea, …
Those files are stored on an SD card, so it's easy to grab them on a computer. (Do not forget to shut down the CPAP before removing the SD card). Here is a sample of the content of such SD card:
.
├── DATALOG
│ ├── 20220302
│ │ ├── 20220303_110420_CSL.crc
│ │ ├── 20220303_110420_CSL.edf
│ │ ├── 20220303_110420_EVE.crc
│ │ ├── 20220303_110420_EVE.edf
│ │ ├── 20220303_110843_BRP.crc
│ │ ├── 20220303_110843_BRP.edf
│ │ ├── 20220303_110844_PLD.crc
│ │ ├── 20220303_110844_PLD.edf
│ │ ├── 20220303_110844_SAD.crc
│ │ └── 20220303_110844_SAD.edf
│ ├── 20220303
│ │ ├── 20220303_234208_CSL.crc
│ │ ├── 20220303_234208_CSL.edf
│ │ ├── 20220303_234208_EVE.crc
│ │ ├── 20220303_234208_EVE.edf
│ │ ├── 20220303_234218_BRP.crc
│ │ ├── 20220303_234218_BRP.edf
│ │ ├── 20220303_234218_PLD.crc
│ │ ├── 20220303_234218_PLD.edf
│ │ ├── 20220303_234218_SAD.crc
│ │ ├── 20220303_234218_SAD.edf
...We developed a tool that converts a single EDF file (or an entire sub-directory, recursively) into GTS format.
$ edf-parser --help
Usage: edf-parser [-hrV] [-d=<directory>] [-f=<file>] [-o=<output>]
[-p=<prefix>]
Parse EDF files
-d, --directory=<directory>
Parse a directory
-f, --file=<file> Parse a file
-h, --help Show this help message and exit.
-o, --output=<output> Output file
-p, --prefix=<prefix> GTS prefix
-r, --recursive Parse a directory recursively
-V, --version Print version information and exit.This tool is (for now) at a beta stage and certainly needs to be improved. If you are interested in using this tool with your files, feel free to contact us.
$ edf-parser -d data/DATALOG -r -p io.senx.edf.cpap -o out.gtsThe result will be something like that:
1647991710689000// io.senx.edf.cpap.Flow.40ms{unit=L%2Fs} 0.43
1647991710729000// io.senx.edf.cpap.Flow.40ms{unit=L%2Fs} 0.418
1647991710769000// io.senx.edf.cpap.Flow.40ms{unit=L%2Fs} 0.41400000000000003
1647991710809000// io.senx.edf.cpap.Flow.40ms{unit=L%2Fs} 0.41000000000000003
1647991710849000// io.senx.edf.cpap.Flow.40ms{unit=L%2Fs} 0.398
1647991710889000// io.senx.edf.cpap.Flow.40ms{unit=L%2Fs} 0.376
1647991710929000// io.senx.edf.cpap.Flow.40ms{unit=L%2Fs} 0.358
1647991710969000// io.senx.edf.cpap.Flow.40ms{unit=L%2Fs} 0.358
1647991711009000// io.senx.edf.cpap.Flow.40ms{unit=L%2Fs} 0.354
...With our test equipment, we have 26 nights of measures and 208 EDF files (62.1 Mb). It produces, a 1.8 Gb gts file containing 30874108 data points (including annotations). I recognize, that our gts text format is not very efficient against the original binary format.
Now we can upload it directly into Warp 10.
$ curl -H 'X-Warp10-Token: TOKEN_WRITE' -H 'Transfer-Encoding: chunked' -T out.gts 'https://HOST:PORT/api/v0/update'Now, you can perform some analysis.
HFile
We recently introduce HFiles. Because we will never update our sleep records, it will be handy to use HFiles to store our data instead of a gts file or by using LevelDB.
You can pipe directly edf-parser with hfadmin. First, create a shell script:
#!/bin/bash
$JAVA_HOME/java \
-Xmx2G \
-Dhfsproducer.license=$LICENCE \
-Dwarp.timeunits=us \
-cp /path/to/warp10/bin/warp10-xxx.jar:/path/to/warp10/lib/warp10-ext-hfsproducer.jar io.senx.hfadmin.Main \
gts \
-a cpap.app \ # Application name
-o d3exxxc1 \ # Owner
-p d3exxxc1 \ # Producer
-c e5xxxc5fc \ # Class hashkeys
-l bcxxxxdbc \ # Labels hashkeys
-g "cpap.gts" \
-i "cpap.info" \
-O "cpap.hfile" \
- # wait for data on the standard input
Do not copy/paste as-this, beware of producer/owner ids, application, and path. Once ready, you can pipe the edf-parser output into your hfile conversion script.
$ edf-parser -d data/DATALOG -r -p io.senx.edf.cpap | ./tohfile.shYou can even shard data by producing a HFile per month for example. Feel free to write a shell script to do so.
The result:
$ du -h cpap.*
8,0K cpap.gts
38M cpap.hfile
4,0K cpap.infoWe still parse our 26 nights of measures and 208 EDF files (62.1 Mb) containing 30874108 data points, but the result fits only in a 38Mb HFile. A datapoint fits approximately in 0,77 byte. Hooray, we can archive, transfer, and use it with a small disk footprint.
Now, you can declare the HFStore in your Warp 10 instance:
...
warp10.plugin.hfstore = io.senx.plugins.hfstore.HFStoreWarp10Plugin
warpscript.extension.hfstore = io.senx.ext.hfstore.HFStoreWarpScriptExtension
plugin.defaultcl.io.senx.plugins.hfstore.HFStoreWarp10Plugin = true
hfstore.secret = 123456
hfstore.stores = cpap # comma separated list of stores
hfstore.root.cpap = /opt/hfstore/cpap
hfstore.secret.cpap = 123456Restart Warp 10 and index it:
{
'secret' '123456'
'store' 'cpap'
'attr' 'store'
'infos' false
'file' 'cpap.gts'
} HFINDEXNow you can access your data efficiently. For instance, on my laptop, I want to read all the datapoints:
{
'store' 'resmed'
'token' $token
'class' '~io.senx.edf.cpap.*'
'labels' {}
'end' MAXLONG
'timespan' MAXLONG
'encoders' false
} HFFETCH DROPYour script execution took 1.962 s serverside, fetched 30873928 datapoints and performed 22 WarpScript operations.In other words, a read rate of 15735947 datapoints per second on my laptop, more than 15 million data per second.
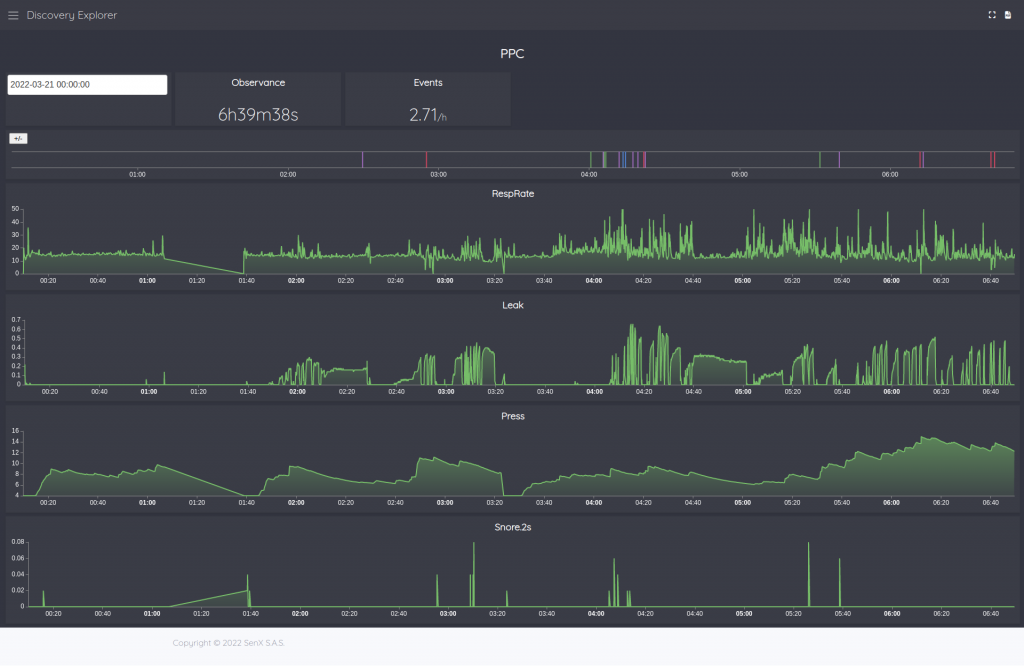
Dashboarding
I will not explain to you how to build a dashboard with Discovery, there are many blog posts for that (Discovery Explorer – The dashboards server, Covid Tracker built with Warp 10 and Discovery, …)
But here is a sample of dashboard:
// @endpoint http://localhost:8080/api/v0/exec
// @theme dark
{
'title' 'CPAP'
'cellHeight' 40
'options' { 'scheme' 'CHARTANA' }
'vars' {
'NOW' NOW
'token' 'owIUxxxxxxxxxxxxxxRGg0Q5.'
}
'tiles' [
{
'type' 'input:date'
'x' 0 'y' 0 'w' 2 'h' 2
'macro' <%
{
'data' $NOW ->TSELEMENTS [ 0 2 ] SUBLIST TSELEMENTS->
'events' [ { 'type' 'variable' 'tags' 'NOW' 'selector' 'NOW' } ]
}
%>
}
{
'type' 'display'
'title' 'Observance'
'options' {
'eventHandler' 'type=variable,tag=NOW'
}
'x' 2 'y' 0 'w' 2 'h' 2
'macro' <%
$NOW ->TSELEMENTS [ 0 2 ] SUBLIST TSELEMENTS-> 12 h + 'end' STORE
{
'store' 'cpap'
'token' $token
'class' 'io.senx.edf.cpap.RespRate.2s'
'labels' {}
'end' $end
'timespan' 24 h
'encoders' false
} HFFETCH 'g' STORE
$g LASTTICK $g FIRSTTICK - HUMANDURATION '.' SPLIT 0 GET 's' +
%>
}
{
'type' 'display'
'title' 'Events'
'unit' '/h'
'options' {
'eventHandler' 'type=variable,tag=NOW'
}
'x' 4 'y' 0 'w' 2 'h' 2
'macro' <%
$NOW ->TSELEMENTS [ 0 2 ] SUBLIST TSELEMENTS-> 12 h + 'end' STORE
{
'store' 'cpap'
'token' $token
'class' 'io.senx.edf.cpap.RespRate.2s'
'labels' {}
'end' $end
'timespan' 24 h
'encoders' false
} HFFETCH 'g' STORE
$g LASTTICK $g FIRSTTICK - 1 h / 1 + 'duration' STORE
{
'store' 'cpap'
'token' $token
'class' 'io.senx.edf.cpap.event'
'labels' { 'desc' '~.*pnea' }
'end' $end
'timespan' 24 h
'encoders' false
} HFFETCH [ SWAP bucketizer.count $g LASTTICK 1 h $duration ] BUCKETIZE
[ NaN NaN NaN 0 ] FILLVALUE
[ SWAP [] reducer.sum ] REDUCE
[ SWAP bucketizer.mean $g LASTTICK 0 1 ] BUCKETIZE VALUES 0 GET 0 GET 100 * ROUND 100.0 /
%>
}
{
'type' 'annotation'
'x' 0 'y' 2 'w' 12 'h' 2
'options' {
'eventHandler' 'type=(variable|zoom|focus),tag=(NOW|chart[2345])'
}
'macro' <%
$NOW ->TSELEMENTS [ 0 2 ] SUBLIST TSELEMENTS-> 12 h + 'end' STORE
{
'store' 'cpap'
'token' $token
'class' 'io.senx.edf.cpap.event'
'labels' { 'desc' '~.*pnea' }
'end' $end
'timespan' 24 h
'encoders' false
} HFFETCH 'data' STORE
{
'store' 'cpap'
'token' $token
'class' 'io.senx.edf.cpap.RespRate.2s'
'labels' {}
'end' $end
'timespan' 24 h
'encoders' false
} HFFETCH 'g' STORE
{
'data' $data
'globalParams' { 'bounds' { 'minDate' $g FIRSTTICK 'maxDate' $g LASTTICK } }
'events' [
{ 'tags' [ 'chart1' ] 'type' 'zoom' }
{ 'tags' [ 'chart1' ] 'type' 'focus' }
]
}
%>
}
{
'type' 'area'
'title' 'RespRate'
'x' 0 'y' 4 'w' 12 'h' 4
'options' {
'eventHandler' 'type=(variable|zoom|focus),tag=(NOW|chart[1345])'
}
'macro' <%
$NOW ->TSELEMENTS [ 0 2 ] SUBLIST TSELEMENTS-> 12 h + 'end' STORE
{
'store' 'cpap'
'token' $token
'class' 'io.senx.edf.cpap.RespRate.2s'
'labels' {}
'end' $end
'timespan' 24 h
'encoders' false
} HFFETCH 'data' STORE
{
'data' $data
'events' [
{ 'tags' [ 'chart2' ] 'type' 'zoom' }
{ 'tags' [ 'chart2' ] 'type' 'focus' }
]
}
%>
}
{
'type' 'area'
'title' 'Leak'
'x' 0 'y' 8 'w' 12 'h' 4
'options' {
'eventHandler' 'type=(variable|zoom|focus),tag=(NOW|chart[1245])'
}
'macro' <%
$NOW ->TSELEMENTS [ 0 2 ] SUBLIST TSELEMENTS-> 12 h + 'end' STORE
{
'store' 'cpap'
'token' $token
'class' 'io.senx.edf.cpap.Leak.2s'
'labels' {}
'end' $end
'timespan' 24 h
'encoders' false
} HFFETCH 'data' STORE
{
'data' $data
'events' [
{ 'tags' [ 'chart3' ] 'type' 'zoom' }
{ 'tags' [ 'chart3' ] 'type' 'focus' }
]
}
%>
}
{
'type' 'area'
'title' 'Press'
'x' 0 'y' 12 'w' 12 'h' 4
'options' {
'eventHandler' 'type=(variable|zoom|focus),tag=(NOW|chart[1235])'
}
'macro' <%
$NOW ->TSELEMENTS [ 0 2 ] SUBLIST TSELEMENTS-> 12 h + 'end' STORE
{
'store' 'cpap'
'token' $token
'class' 'io.senx.edf.cpap.Press.2s'
'labels' {}
'end' $end
'timespan' 24 h
'encoders' false
} HFFETCH 'data' STORE
{
'data' $data
'events' [
{ 'tags' [ 'chart4' ] 'type' 'zoom' }
{ 'tags' [ 'chart4' ] 'type' 'focus' }
]
}
%>
}
{
'type' 'area'
'title' 'Snore.2s'
'x' 0 'y' 16 'w' 12 'h' 4
'options' {
'eventHandler' 'type=(variable|zoom|focus),tag=(NOW|chart[1234])'
}
'macro' <%
$NOW ->TSELEMENTS [ 0 2 ] SUBLIST TSELEMENTS-> 12 h + 'end' STORE
{
'store' 'cpap'
'token' $token
'class' 'io.senx.edf.cpap.Snore.2s'
'labels' {}
'end' $end
'timespan' 24 h
'encoders' false
} HFFETCH 'data' STORE
{
'data' $data
'events' [
{ 'tags' [ 'chart5' ] 'type' 'zoom' }
{ 'tags' [ 'chart5' ] 'type' 'focus' }
]
}
%>
}
]
}Going further
You should shard your HFiles depending on your dataset and your use cases.
Wants to learn more about EDF parser and HFiles?
Read more
Truly Dynamic Dashboards as Code
Demystifying the use of the Parquet file format for time series
Grafana BeerTender Dashboard connected with Warp 10

Senior Software Engineer